Project description
Project description
The challenge: to make ecology a predictive science
One of the grand challenges in ecology is to understand and predict how ecosystems are impacted by changes in environmental conditions, and external pressures. The urgency of this challenge is high given the unprecedented rate of global change in climate, land use, and urbanisation. To do so, we need to transform ecology into a predictive science. Scenarios can be used to explore how complex ecosystems could behave under different external pressures. Developing scenarios requires combining knowledge and data. This is analogous to global climate models, which also integrate fundamental knowledge and empirical data on climate processes to forecast consequences of different global emission scenarios. Key for developing high-quality scenarios is the availability of long-term environmental and ecological datasets. These will become available in a user-friendly and secure cloud-based digital modelling and simulation platform. This platform can also be used to link this data to models. The Netherlands, with its long tradition of long-term data collection on plants and animals, and modelling of ecological processes, is the perfect place to do this.
The approach: Digital Twins of entire ecosystems
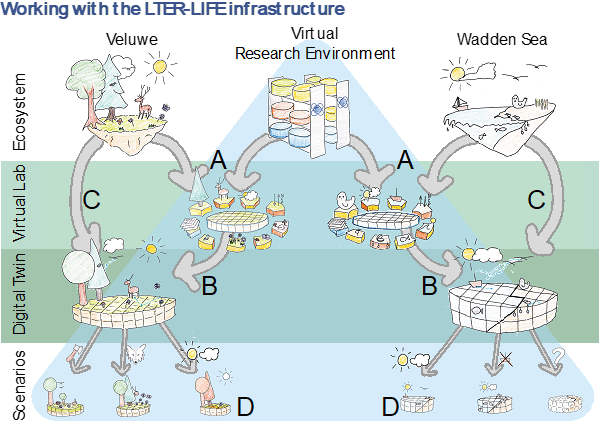
A Digital Twin is “a digital replica of a living or non-living physical entity”. Digital Twins allow advanced data-driven modelling and simulation, using Big Data tools to generate novel insights that cannot be obtained with traditional observation models. Building Digital Twins of ecosystems has only recently became possible as Big Data, artificial intelligence (AI) applications and analytics, advanced computing infrastructure, and the FAIR principles have been developed and made available for ecology, ecosystem restoration, and biodiversity science. A Digital Twin of an ecosystem also provides tools to integrate data on abiotic factors (e.g., nutrient deposition, temperature, droughts), biotic factors (e.g., long-term occurrence data on animals and plants, life history data), and human activities (e.g., tourism, agriculture, fishery). It provides diagnostic (data-driven) and dynamic (process-based) models in one logical place, ensuring their necessary interoperability, scalability, storage, and processing capacity (Figure 1). Thus, ‘‘Big Data’’ collected on ecosystems and model-based data integration can be coupled in an unprecedented way with well-developed process-based models on the relationships between species and their environment. This approach brings together data scientists, informaticians, and ecologists in research communities around ecosystems.
The infrastructure: LTER-LIFE
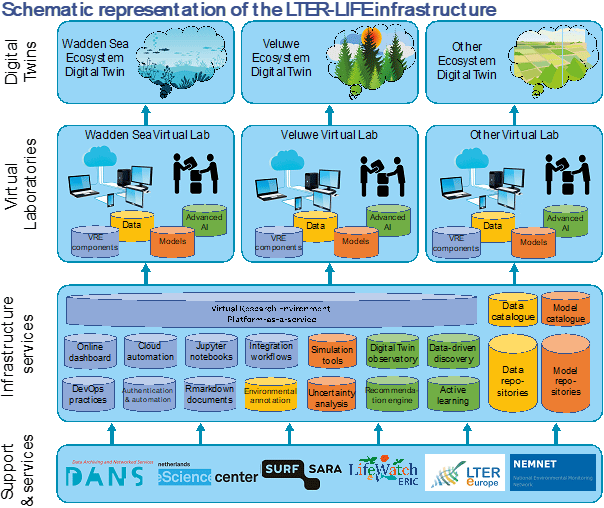
To produce Digital Twins of entire ecosystems, LTER-LIFE will develop an infrastructure that includes a Virtual Research Environment (VRE) and services (e.g., catalogues and repositories that contain FAIR data, models, and software tools) that together can instantiate custom Virtual Laboratories to build Digital Twins of specific ecosystems (Figure 2 ).
Virtual laboratories
The Virtual Laboratories provide VRE components, data, models, and other Big Data tools (e.g., AI) and allow scientific users to discover, access, and integrate research assets for specific scientific purposes, and manage the deployment, execution, and provenance of in silico experiments (analysis and simulations) on local or remote platforms. This includes the following assets:
- FAIR datasets of long-term studies of plants and animal populations as well as of environmental data. These data can be (a) at the data owner when FAIR and open, (b) at an LTER-LIFE server if the data owner cannot facilitate this, or (c) managed elsewhere (data or model outputs from authoritative sources such as remote-sensing observations, climate model projections or historical weather data).
- FAIR models such as (a) process-based models that formalise ecological knowledge on abiotic and biotic relationships, or (b) data-driven models that explore relationships between data using classical statistics, or more contemporary artificial intelligent tools. These models themselves are made FAIR, so that they can be discovered, accessed, and reused in new scientific workflows.
- Rules of interactions between assets (i.e., data and models), including solutions for scaling and enhancing interoperability of datasets, which are collected with different methodology.
- Tools for scenario studies to explore the effects of in situ local management strategies on ecosystem functioning.
These assets can be coupled in various ways, enabled by the LTER-LIFE platform depending on the objectives of the specific research. Together, they will ultimately compose a Digital Twin of a specific ecosystem, which can then be used for scientific research or scenario studies to address societal issues.
Summarizing
With LTER-LIFE, we will offer a single point of entry for scientists studying entire ecosystems or components thereof. The LTER-LIFE portal gives users tailored access to assets and facilities. LTER-LIFE develops:
- Data bases and process models that are findable, accessible, interoperable, and reusable (FAIR, by applying (meta)data standards, controlled vocabularies, ontologies, and persistent identifiers).
- Workflows that allow coupling of these data and models at the desired temporal and spatial scale, and that provide Big Data methodology to analyse these data (AI and machine learning approaches such as deep learning and active learning which enable new theory discovery and novel insights from high-dimensional sparse data spaces) as well as tools for uncertainty analysis, scenario studies and forecasting.
- Virtual Labs that build upon infrastructure services and components of a VRE, allowing interdisciplinary communities of scientists to collaborate via a digital platform (including Big Data storage, cloud-based modelling, and dynamic simulations).
- Basic and advanced training in the use of the infrastructure.
LTER-LIFE will be an open-source infrastructure and users can either use the developed assets within their own local environment or run it in the cloud, facilitated by LTER-LIFE through our partner SURF. Setting up the LSRI LTER-LIFE as an open-source infrastructure aligns well with the current FAIR and Open Science developments. New data or models can be added as assets, which can then be used by other users. Workflows and data as used in projects will receive a digital identifier and will be stored by LTER-LIFE’s partners DANS-KNAW and SURF.